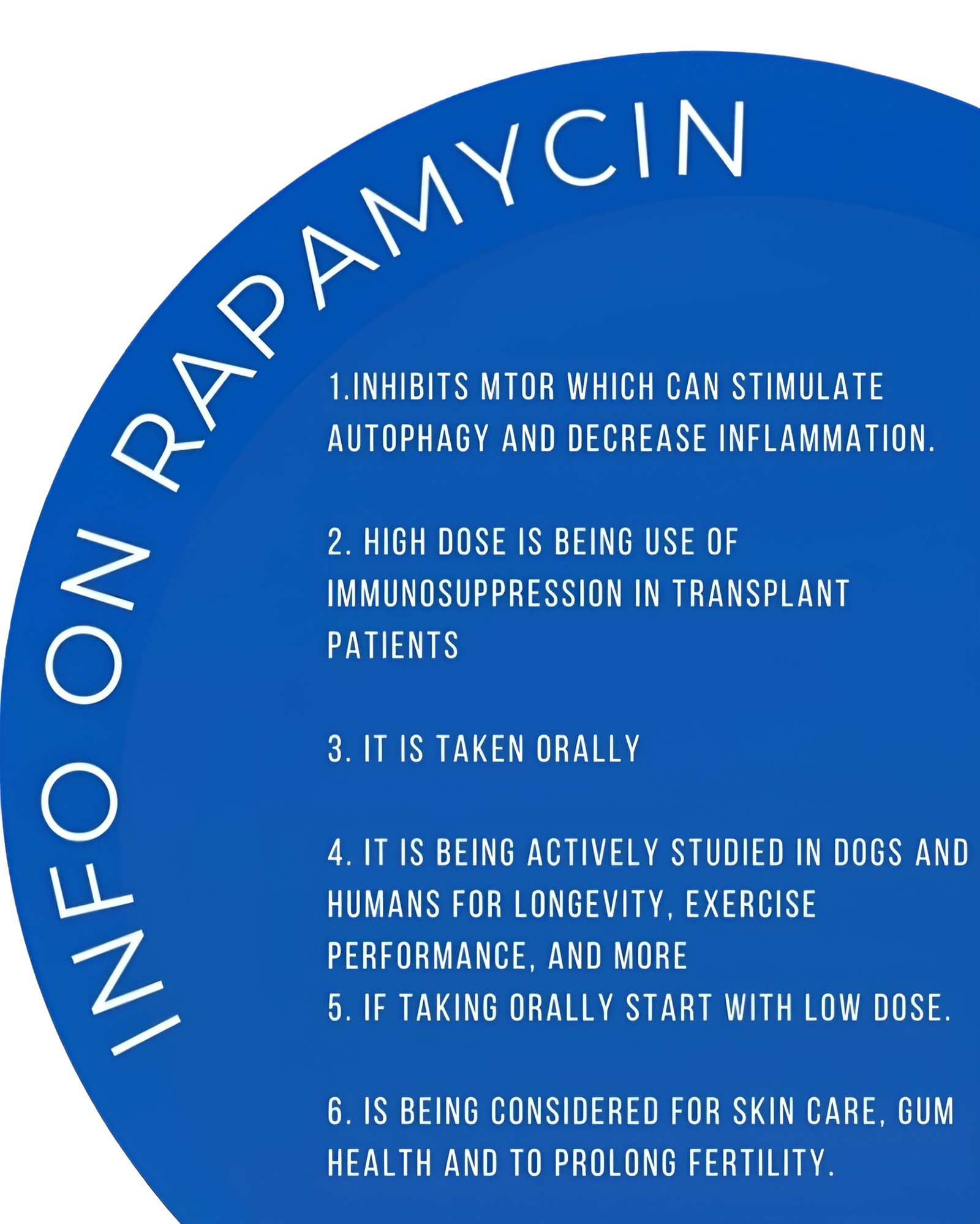
In the realm of cellular biology, the process of autophagy has emerged as a key player in maintaining cellular health and homeostasis. It’s like the cell’s own spring cleaning mechanism, where damaged or unnecessary components are broken down and recycled to ensure optimal functioning. In recent years, the discovery of rapamycin-induced autophagy has sparked immense interest in the scientific community, paving the way for potential therapeutic interventions in various diseases, including cancer, neurodegenerative disorders, and aging.
Understanding Autophagy: A Cellular Cleanup Crew
Before delving into rapamycin-induced autophagy, let’s grasp the fundamentals of autophagy itself. Autophagy, derived from the Greek words “auto” (self) and “phagy” (eating), is a highly conserved cellular process essential for maintaining cellular health and survival. It involves the degradation and recycling of damaged organelles, misfolded proteins, and other cellular components through the lysosomal machinery. This self-cleaning mechanism not only ensures cellular renewal but also plays a crucial role in adapting to various stressors such as nutrient deprivation, oxidative stress, and infection.
Rapamycin: From Antibiotic to Autophagy Inducer
Rapamycin, initially discovered as a natural product produced by the bacterium Streptomyces hygroscopicus found in the soil of Easter Island (Rapa Nui), gained attention for its potent immunosuppressive properties. It inhibits the mechanistic target of rapamycin (mTOR), a central regulator of cell growth, proliferation, and metabolism. By inhibiting mTOR, rapamycin induces autophagy, thereby promoting cellular recycling and survival.
The Mechanisms Behind Rapamycin-Induced Autophagy
The mTOR pathway exists in two complexes: mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2). While mTORC1 inhibition by rapamycin is well-established as a trigger for autophagy, recent research has revealed intricate crosstalk and feedback mechanisms between mTOR complexes, autophagy, and other cellular processes. These findings highlight the complexity of rapamycin-induced autophagy and underscore the need for further exploration to unravel its full therapeutic potential.
Potential Therapeutic Applications
The discovery of rapamycin-induced autophagy has opened new avenues for therapeutic interventions in various diseases. In cancer, where dysregulated mTOR signaling is commonly observed, rapamycin and its analogs (rapalogs) hold promise as adjuvant therapies to enhance the efficacy of conventional treatments and overcome resistance mechanisms. Additionally, rapamycin-induced autophagy shows potential in combating neurodegenerative disorders such as Alzheimer’s and Parkinson’s disease by clearing toxic protein aggregates implicated in disease pathology.
Challenges and Future Directions
Despite its therapeutic potential, rapamycin and rapalogs face challenges such as off-target effects, dose-limiting toxicity, and the complexity of autophagy regulation. Moreover, the dual role of autophagy in cancer, acting as both a tumor suppressor and a pro-survival mechanism, necessitates careful consideration in therapeutic strategies.
Future research directions include elucidating the specific molecular mechanisms underlying rapamycin-induced autophagy, developing targeted delivery systems to minimize off-target effects, and exploring combination therapies to enhance efficacy and minimize resistance.
Conclusion
Rapamycin-induced autophagy represents a promising avenue for therapeutic intervention in various diseases by harnessing the cell’s innate ability to maintain homeostasis and combat stress. As research continues to unravel the complexities of autophagy regulation and mTOR signaling, the potential clinical applications of rapamycin and its derivatives are poised to revolutionize medicine and pave the way for personalized therapies tailored to individual patient needs.